High diversity in the TCR repertoire of GAD65 autoantigen-specific human CD4+ T cells.
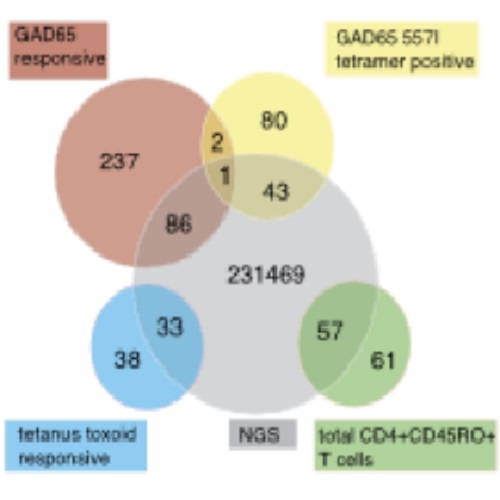
Autoreactive CD4(+) T cells are an essential feature of type 1 diabetes mellitus. We applied single-cell TCR ?- and ?-chain sequencing to peripheral blood GAD65-specific CD4(+) T cells, and TCR ?-chain next-generation sequencing to bulk memory CD4(+) T cells to provide insight into TCR diversity in autoimmune diabetes mellitus. TCRs obtained for 1650 GAD65-specific CD4(+) T cells isolated from GAD65 proliferation assays and/or GAD65 557I tetramer staining in 6 patients and 10 islet autoantibody-positive children showed large diversity with 1003 different TCRs identified. TRAV and TRBV gene usage was broad, and the TRBV5.1 gene was most prominent within the GAD65 557I tetramer(+) cells. Limited overlap (<5%) was observed between TCRs of GAD65-proliferating and GAD65 557I tetramer(+) CD4(+) T cells. Few TCRs were repeatedly found in GAD65-specific cells at different time points from individual patients, and none was seen in more than one subject. However, single chains were often shared between patients and used in combination with different second chains. Next-generation sequencing revealed a wide frequency range (<0.00001-1.62%) of TCR ?-chains corresponding to GAD65-specific T cells. The findings support minor selection of genes and TCRs for GAD65-specific T cells, but fail to provide strong support for TCR-targeted therapies.
- J. Immunol. 2015 Mar 15;194(6):2531-8
- 2015
- Medical Biology
- 25681349
- PubMed
Enabled by:
