Correlative single-molecule localization microscopy and electron tomography reveals endosome nanoscale domains.
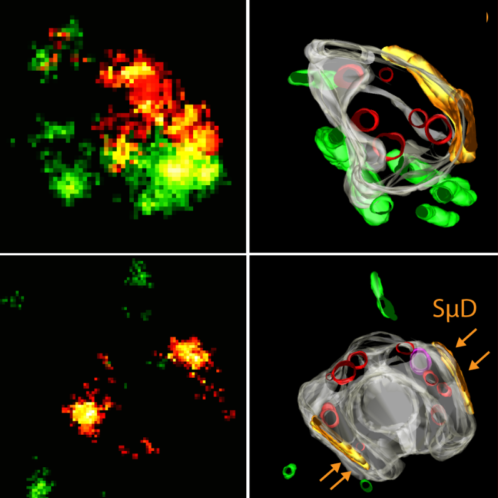
Many cellular organelles, including endosomes, show compartmentalization into distinct functional domains, which, however, cannot be resolved by diffraction-limited light microscopy. Single molecule localization microscopy (SMLM) offers nanoscale resolution but data interpretation is often inconclusive when the ultrastructural context is missing. Correlative light electron microscopy (CLEM) combining SMLM with electron microscopy (EM) enables correlation of functional subdomains of organelles in relation to their underlying ultrastructure at nanometer resolution. However, the specific demands for EM sample preparation and the requirements for fluorescent single-molecule photo-switching are opposed. Here, we developed a novel superCLEM workflow that combines triple-color SMLM (dSTORM & PALM) and electron tomography using semi-thin Tokuyasu thawed cryosections. We applied the superCLEM approach to directly visualize nanoscale compartmentalization of endosomes in HeLa cells. Internalized, fluorescently labeled Transferrin and EGF were resolved into morphologically distinct domains within the same endosome. We found that the small GTPase Rab5 is organized in nanodomains on the globular part of early endosomes. The simultaneous visualization of several proteins in functionally distinct endosomal sub-compartments demonstrates the potential of superCLEM to link the ultrastructure of organelles with their molecular organization at nanoscale resolution.
- Traffic. 2019 Aug;20(8):601-617
- 2019
- Imaging Technologies Development
- 31206952
- PubMed
Enabled by:
